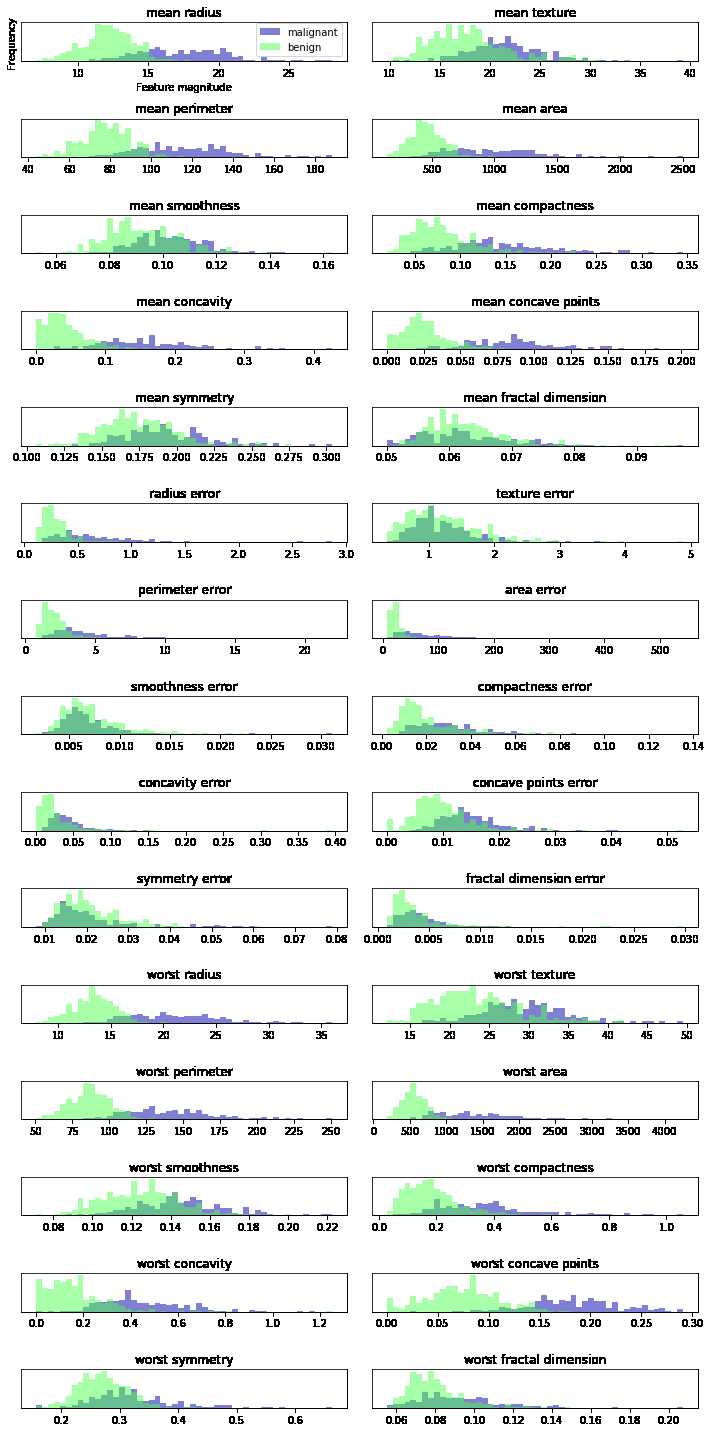
import graphviz import mglearn from mpl_toolkits.mplot3d import Axes3D from sklearn.datasets import load_breast_cancer, make_blobs from sklearn.ensemble import RandomForestClassifier from sklearn.model_selection import train_test_split from sklearn.svm import SVC from sklearn.tree import DecisionTreeClassifier, export_graphviz from IPython.display import display import matplotlib.pyplot as plt import numpy as np import matplotlib as mt import pandas as pd from sklearn.datasets import load_breast_cancer from sklearn.model_selection import train_test_split cancer = load_breast_cancer() # X_train, X_test, y_train, y_test = train_test_split(cancer.data, cancer.target, # random_state=1) fig, axes = plt.subplots(15, 2, figsize=(10, 20)) malignant = cancer.data[cancer.target == 0] benign = cancer.data[cancer.target == 1] ax = axes.ravel() # 直方图显示了数据值的分布情况 for i in range(30): _, bins = np.histogram(cancer.data[:, i], bins=50) # 逐列取数 ax[i].hist(malignant[:, i], bins=bins, color=mglearn.cm3(0), alpha=.5) ax[i].hist(benign[:, i], bins=bins, color=mglearn.cm3(2), alpha=.5) ax[i].set_title(cancer.feature_names[i]) ax[i].set_yticks(()) ax[0].set_xlabel("Feature magnitude") ax[0].set_ylabel("Frequency") ax[0].legend(["malignant", "benign"], loc="best") fig.tight_layout() plt.show()
原文:https://www.cnblogs.com/starcrm/p/11686898.html