紧接上文: 计算二维离散随机变量的联合概率分布
我们知道了上文提到的几种计算二维概率密度分布中, accumarray 方法是最快的.
那么就使用accumarray来求计算两幅相同大小图像的归一化互信息.
离散变量的互信息定义为:

求联合分布和边缘分布会用到了上文的方法.
或者使用熵来定义:
其中,
归一化互信息定义为:
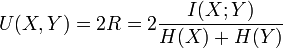
所以, 不论是求互信息还是求归一化互信息, 都要把两个随机变量的联合分布和边缘分布求出来.
而边缘分布可以从联合分布求出来. 所以只要求出来联合分布就可以了. 这正是我们上文的主题!
求互信息和归一化互信息的函数, 在子桥的cnblogs 基础上修改:
function [MI,NMI] = NormMutualInfo( A, B ,method)
% NMI Normalized mutual information
% http://en.wikipedia.org/wiki/Mutual_information
% http://nlp.stanford.edu/IR-book/html/htmledition/evaluation-of-clustering-1.html
% % Example :
% % (http://nlp.stanford.edu/IR-book/html/htmledition/evaluation-of-clustering-1.html)
% % A = [1 1 1 1 1 1 2 2 2 2 2 2 3 3 3 3 3];
% % B = [1 2 1 1 1 1 1 2 2 2 2 3 1 1 3 3 3];
% % nmi(A,B)
%
% % ans = 0.3646
switch method
case 1
% 参考http://www.cnblogs.com/ziqiao/archive/2011/12/13/2286273.html#3350670
if length( A ) ~= length( B)
error(‘length( A ) must == length( B)‘);
end
total = length(A);
A_ids = unique(A);
B_ids = unique(B);
% Mutual information
MI = 0;
for idA = A_ids
for idB = B_ids
idAOccur = find( A == idA );
idBOccur = find( B == idB );
idABOccur = intersect(idAOccur,idBOccur);
px = length(idAOccur)/total;
py = length(idBOccur)/total;
pxy = length(idABOccur)/total;
MI = MI + pxy*log2(pxy/(px*py)+eps); % eps : the smallest positive number
end
end
% Normalized Mutual information
Hx = 0; % Entropies
for idA = A_ids
idAOccurCount = length( find( A == idA ) );
Hx = Hx - (idAOccurCount/total) * log2(idAOccurCount/total + eps);
end
Hy = 0; % Entropies
for idB = B_ids
idBOccurCount = length( find( B == idB ) );
Hy = Hy - (idBOccurCount/total) * log2(idBOccurCount/total + eps);
end
NMI = 2 * MI / (Hx+Hy);
case 2
% 使用accumarray方法
A=A(:);
B=B(:);
if length(A)~=length(B)
error(‘A B must be the SAME length!\n‘);
end
H=accumarray([A B],ones(1,size(A,1)));
Pab=H/length(A);
pa=sum(Pab,2);
pb=sum(Pab,1);
Pa=repmat(pa,1,size(Pab,2));
Pb=repmat(pb,size(Pab,1),1);
MI=sum(sum(Pab.*log((Pab+eps)./(Pa.*Pb+eps)+eps)));
Ha=-sum(pa.*log(pa+eps));% 熵
Hb=-sum(pb.*log(pb+eps));
NMI=2*MI/(Ha+Hb);
end
end
测试脚本:
A = randi(256,1,1e3);
B = randi(256,1,1e3);
tic
[mi1,nmi1]=NormMutualInfo(A,B,1)
toc
tic
[mi2,nmi2]=NormMutualInfo(A,B,2)
toc输出:
mi1 =
5.6650
nmi1 =
0.7257
Elapsed time is 4.862238 seconds.
mi2 =
3.9267
nmi2 =
0.7257
Elapsed time is 0.003858 seconds.NMI相同, 但是MI不相同.
多试几次, 基本上method2 比method1快了1000多倍!
类似matlab中的normxcorr2函数, 如果输入两个矩阵的大小
为了节省不必要的计算. NMI矩阵大小为
NMI(1,1)代表两个矩阵左上角(1,1)元素对齐求出的NMI. NMI(end,end)代表两个矩阵右下角对齐.
method1, 为了进一步节省计算量, template矩阵的边缘分布和熵事先求出.
method2, 直接使用相同大小的矩阵调用accumarray.
matlab函数
function [ MI,NMI] = MI_matrix( A,B,L,method )
switch method
case 1
% 求NMI矩阵, 事先求出template边缘分布Pb和熵Hb
[M,N]=size(A);
[m,n]=size(B);
pb=hist(B(:),1:L)/(m*n);
Hb=-sum(pb.*log(pb+eps));
Pb=repmat(pb,L,1);
MI=nan(M-m+1,N-n+1);
NMI=nan(M-m+1,N-n+1);
for i=1:M-m+1
for j=1:N-n+1
ImgSub=A(i:i+m-1,j:j+n-1);
H=accumarray([ImgSub(:) B(:)],ones(1,m*n));
Pab=H/(m*n);
pa=sum(Pab,2);
Pa=repmat(pa,1,size(Pab,2));
MI(i,j)=sum(sum(Pab.*log((Pab+eps)./(Pa.*Pb+eps)+eps)));
Ha=-sum(pa.*log(pa+eps));% 熵
NMI(i,j)=2*MI(i,j)/(Ha+Hb);
end
end
case 2
% 求NMI矩阵
[M,N]=size(A);
[m,n]=size(B);
MI=nan(M-m+1,N-n+1);
NMI=nan(M-m+1,N-n+1);
for i=1:M-m+1
for j=1:N-n+1
ImgSub=A(i:i+m-1,j:j+n-1);
H=accumarray([ImgSub(:) B(:)],ones(1,m*n));
Pab=H/(m*n);
pa=sum(Pab,2);
pb=sum(Pab,1);
Pa=repmat(pa,1,size(Pab,2));
Pb=repmat(pb,size(Pab,1),1);
MI(i,j)=sum(sum(Pab.*log((Pab+eps)./(Pa.*Pb+eps)+eps)));
Ha=-sum(pa.*log(pa+eps));% 熵
Hb=-sum(pb.*log(pb+eps));% 熵
NMI(i,j)=2*MI(i,j)/(Ha+Hb);
end
end
end测试代码:
A = randi(256,256,256);%256*256大小
B = randi(256,200,200);%200*200大小
tic
[ MI1,NMI1] = MI_matrix( A,B,256,1);
toc
tic
[ MI2,NMI2] = MI_matrix( A,B,256,2);
toc
all(abs(MI1(:)-MI2(:))<1e-5)
all(abs(NMI1(:)-NMI2(:))<1e-5)输出:
Elapsed time is 7.564679 seconds.
Elapsed time is 8.167062 seconds.
ans =
1
ans =
1实验表明, A和B大小相差越大, method1节省时间的优势越大.
如果A和B大小相差不大, method2更快一些.
图像的归一化互信息Normlized Mutual Information
原文:http://blog.csdn.net/full_speed_turbo/article/details/69566101