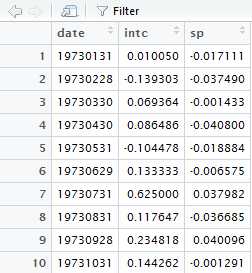
intc=log(da$intc+1)intc [1] 0.0099998346-0.1500127529 0.0670640792 [4] 0.0829486352-0.1103484905 0.1251628488 [7] 0.4855078158 0.1112255825 0.2109235908..........library(fGarch)da=read.table("m-intcsp7309.txt",header=T)intc=log(da$intc+1)m4=garchFit(~1+garch(1,1),data=intc)summary(m4)Title: GARCH Modelling Call: garchFit(formula =~1+ garch(1,1), data = intc) MeanandVarianceEquation: data ~1+ garch(1,1)<environment:0x000000001d475658> [data = intc]ConditionalDistribution: norm Coefficient(s): mu omega alpha1 beta1 0.01126568 0.00091902 0.08643831 0.85258554 Std.Errors: based on Hessian ErrorAnalysis: Estimate Std.Error t value Pr(>|t|) mu 0.0112657 0.0053931 2.089 0.03672* omega 0.0009190 0.0003888 2.364 0.01808* alpha1 0.0864383 0.0265439 3.256 0.00113** beta1 0.8525855 0.0394322 21.622 <2e-16***---Signif. codes: 0‘***’0.001‘**’0.01‘*’0.05‘.’0.1‘’1LogLikelihood: 312.3307 normalized: 0.7034475 Description: FriJan2721:49:062017 by user: xuan StandardisedResidualsTests: Statistic p-Value Jarque-BeraTest R Chi^2 174.904 0 Shapiro-WilkTest R W 0.97096151.030282e-07 Ljung-BoxTest R Q(10) 8.016844 0.6271916 Ljung-BoxTest R Q(15) 15.5006 0.4159946 Ljung-BoxTest R Q(20) 16.41549 0.6905368 Ljung-BoxTest R^2 Q(10) 0.87463450.9999072 Ljung-BoxTest R^2 Q(15) 11.35935 0.7267295 Ljung-BoxTest R^2 Q(20) 12.55994 0.8954573 LM ArchTest R TR^2 10.51401 0.5709617 InformationCriterionStatistics: AIC BIC SIC HQIC -1.388877-1.351978-1.389037-1.374326 function (formula =~garch(1,1), data = dem2gbp, init.rec = c("mci", "uev"), delta =2, skew =1, shape =4, cond.dist = c("norm", "snorm","ged","sged","std","sstd","snig","QMLE"), include.mean = TRUE, include.delta = NULL, include.skew = NULL, include.shape = NULL, leverage = NULL, trace = TRUE, algorithm = c("nlminb", "lbfgsb","nlminb+nm","lbfgsb+nm"), hessian = c("ropt", "rcd"), control = list(), title = NULL, description = NULL, ...) { DEBUG = FALSE init.rec = match.arg(init.rec) cond.dist = match.arg(cond.dist) hessian = match.arg(hessian) algorithm = match.arg(algorithm) CALL = match.call() Name= capture.output(substitute(data)) if(is.character(data)){ eval(parse(text = paste("data(", data,")"))) data = eval(parse(text = data)) } data <-as.data.frame(data) if(isUnivariate(data)){ colnames(data)<-"data" } else{ uniqueNames = unique(sort(colnames(data))) if(is.null(colnames(data))){ stop("Column names of data are missing.") } if(length(colnames(data))!= length(uniqueNames)){ stop("Column names of data are not unique.") } } if(length(formula)==3&& isUnivariate(data)) formula[2]<- NULL if(length(formula)==2){ if(isUnivariate(data)){ formula =as.formula(paste("data", paste(formula, collapse =" "))) } else{ stop("Multivariate data inputs require lhs for the formula.") } } robust.cvar <-(cond.dist =="QMLE") args =.garchArgsParser(formula = formula, data = data, trace = FALSE) if(DEBUG) print(list(formula.mean = args$formula.mean, formula.var = args$formula.var, series = args$series, init.rec = init.rec, delta = delta, skew = skew, shape = shape, cond.dist = cond.dist, include.mean = include.mean, include.delta = include.delta, include.skew = include.skew, include.shape = include.shape, leverage = leverage, trace = trace, algorithm = algorithm, hessian = hessian, robust.cvar = robust.cvar, control = control, title = title, description = description)) ans =.garchFit(formula.mean = args$formula.mean, formula.var = args$formula.var, series = args$series, init.rec, delta, skew, shape, cond.dist, include.mean, include.delta, include.skew, include.shape, leverage, trace, algorithm, hessian, robust.cvar, control, title, description,...) ans@call = CALL attr(formula,"data")<- paste("data = ",Name, sep ="") ans@formula = formula ans} init.rec = match.arg(init.rec) cond.dist = match.arg(cond.dist) hessian = match.arg(hessian) algorithm = match.arg(algorithm) CALL = match.call()formal.args <- formals(sys.function(sys.parent())).......................................if(identical(arg, choices)) return(arg[1L]) 跳出函数
跳出函数
args =.garchArgsParser(formula = formula, data = data, trace = FALSE)function (formula, data, trace = FALSE){ allVars = unique(sort(all.vars(formula))) allVarsTest = mean(allVars %in% colnames(data))if(allVarsTest !=1){print(allVars)print(colnames(data)) stop("Formula and data units do not match.")} formula.lhs =as.character(formula)[2] mf = match.call(expand.dots = FALSE)if(trace){ cat("\nMatched Function Call:\n ")print(mf)} m = match(c("formula","data"), names(mf),0) mf = mf[c(1, m)] mf[[1]]=as.name(".garchModelSeries") mf$fake = FALSE mf$lhs = TRUEif(trace){ cat("\nModelSeries Call:\n ")print(mf)} x = eval(mf, parent.frame())if(trace)print(x) x =as.vector(x[,1]) names(x)= rownames(data)if(trace)print(x) allLabels = attr(terms(formula),"term.labels")if(trace){ cat("\nAll Term Labels:\n ")print(allLabels)}if(length(allLabels)==2){ formula.mean =as.formula(paste("~", allLabels[1])) formula.var =as.formula(paste("~", allLabels[2]))}elseif(length(allLabels)==1){ formula.mean =as.formula("~ arma(0, 0)") formula.var =as.formula(paste("~", allLabels[1]))}if(trace){ cat("\nMean Formula:\n ")print(formula.mean) cat("\nVariance Formula:\n ")print(formula.var)} ans <- list(formula.mean = formula.mean, formula.var = formula.var, formula.lhs = formula.lhs, series = x) ans}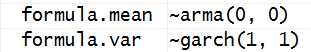 (方程的基本形式已经被解析出来了)
(方程的基本形式已经被解析出来了)
ans =.garchFit(formula.mean = args$formula.mean, formula.var = args$formula.var, series = args$series, init.rec, delta, skew, shape, cond.dist, include.mean, include.delta, include.skew, include.shape, leverage, trace, algorithm, hessian, robust.cvar, control, title, description,...)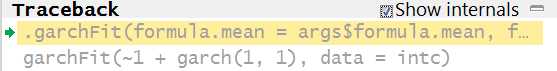
 可以选择环境,一般情况是在global environment
可以选择环境,一般情况是在global environmentfunction (formula.mean =~arma(0,0), formula.var =~garch(1,1), series, init.rec = c("mci","uev"), delta =2, skew =1, shape =4, cond.dist = c("norm","snorm","ged","sged","std","sstd","QMLE"), include.mean = TRUE, include.delta = NULL, include.skew = NULL, include.shape = NULL, leverage = NULL, trace = TRUE, algorithm = c("sqp","nlminb","lbfgsb","nlminb+nm","lbfgsb+nm"), hessian = c("ropt","rcd"), robust.cvar, control = list(), title = NULL, description = NULL,...){ DEBUG <- FALSEif(DEBUG)print("Formula Specification ...") fcheck = rev(all.names(formula.mean))[1]if(fcheck =="ma"){ stop("Use full formula: arma(0,q) for ma(q)")}elseif(fcheck =="ar"){ stop("Use full formula expression: arma(p,0) for ar(p)")}if(DEBUG)print("Recursion Initialization ...")if(init.rec[1]!="mci"& algorithm[1]!="sqp"){ stop("Algorithm only supported for mci Recursion")}.StartFit<-Sys.time()if(DEBUG)print("Generate Control List ...") con <-.garchOptimizerControl(algorithm, cond.dist) con[(namc <- names(control))]<- controlif(DEBUG)print("Initialize Time Series ...") data <- series scale <-if(con$xscale) sd(series)else1 series <- series/scale.series <-.garchInitSeries(formula.mean = formula.mean, formula.var = formula.var, cond.dist = cond.dist[1], series = series, scale = scale, init.rec = init.rec[1], h.start = NULL, llh.start = NULL, trace = trace).setfGarchEnv(.series =.series)if(DEBUG)print("Initialize Model Parameters ...").params <-.garchInitParameters(formula.mean = formula.mean, formula.var = formula.var, delta = delta, skew = skew, shape = shape, cond.dist = cond.dist[1], include.mean = include.mean, include.delta = include.delta, include.skew = include.skew, include.shape = include.shape, leverage = leverage, algorithm = algorithm[1], control = con, trace = trace).setfGarchEnv(.params =.params)if(DEBUG)print("Select Conditional Distribution ...").setfGarchEnv(.garchDist =.garchSetCondDist(cond.dist[1]))if(DEBUG)print("Estimate Model Parameters ...").setfGarchEnv(.llh =1e+99).llh <-.getfGarchEnv(".llh") fit =.garchOptimizeLLH(hessian, robust.cvar, trace)if(DEBUG)print("Add to fit ...").series <-.getfGarchEnv(".series").params <-.getfGarchEnv(".params") names(.series$h)<- NULL fit$series =.series fit$params =.paramsif(DEBUG)print("Retrieve Residuals and Fitted Values ...") residuals =.series$z fitted.values =.series$x - residuals h.t =.series$hif(.params$includes["delta"]) deltainv =1/fit$par["delta"]else deltainv =1/fit$params$delta sigma.t =(.series$h)^deltainvif(DEBUG)print("Standard Errors and t-Values ...") fit$cvar <-if(robust.cvar)(solve(fit$hessian)%*%(t(fit$gradient)%*% fit$gradient)%*% solve(fit$hessian))else-solve(fit$hessian) fit$se.coef = sqrt(diag(fit$cvar)) fit$tval = fit$coef/fit$se.coef fit$matcoef = cbind(fit$coef, fit$se.coef, fit$tval,2*(1- pnorm(abs(fit$tval)))) dimnames(fit$matcoef)= list(names(fit$tval), c(" Estimate"," Std. Error"," t value","Pr(>|t|)"))if(DEBUG)print("Add Title and Description ...")if(is.null(title)) title ="GARCH Modelling"if(is.null(description)) description = description()Time=Sys.time()-.StartFitif(trace){ cat("\nTime to Estimate Parameters:\n ")print(Time)} new("fGARCH", call =as.call(match.call()), formula =as.formula(paste("~", formula.mean,"+", formula.var)), method ="Max Log-Likelihood Estimation", data = data, fit = fit, residuals = residuals, fitted = fitted.values, h.t = h.t, sigma.t =as.vector(sigma.t), title =as.character(title), description =as.character(description))}scale <-if(con$xscale) sd(series)series <- series/scale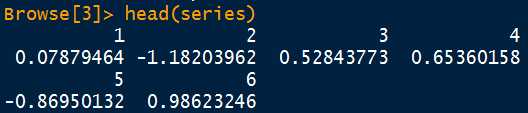
.series <-.garchInitSeries(formula.mean = formula.mean, formula.var = formula.var, cond.dist = cond.dist[1], series = series, scale = scale, init.rec = init.rec[1], h.start = NULL, llh.start = NULL, trace = trace).setfGarchEnv(.series =.series)function (formula.mean, formula.var, cond.dist, series, scale, init.rec, h.start, llh.start, trace) { mm = length(formula.mean) if(mm !=2) stop("Mean Formula misspecified") end = regexpr("\\(",as.character(formula.mean[mm]))-1 model.mean = substr(as.character(formula.mean[mm]),1, end) if(!any(c("ar","ma","arma")== model.mean)) stop("formula.mean must be one of: ar, ma, arma") mv = length(formula.var) if(mv !=2) stop("Variance Formula misspecified") end = regexpr("\\(",as.character(formula.var[mv]))-1 model.var = substr(as.character(formula.var[mv]),1, end) if(!any(c("garch","aparch")== model.var)) stop("formula.var must be one of: garch, aparch") model.order =as.numeric(strsplit(strsplit(strsplit(as.character(formula.mean), "\\(")[[2]][2],"\\)")[[1]],",")[[1]]) u = model.order[1] v =0 if(length(model.order)==2) v = model.order[2] maxuv = max(u, v) if(u <0| v <0) stop("*** ARMA orders must be positive.") model.order =as.numeric(strsplit(strsplit(strsplit(as.character(formula.var), "\\(")[[2]][2],"\\)")[[1]],",")[[1]]) p = model.order[1] q =0 if(length(model.order)==2) q = model.order[2] if(p + q ==0) stop("Misspecified GARCH Model: Both Orders are zero!") maxpq = max(p, q) if(p <0| q <0) stop("*** GARCH orders must be positive.") max.order = max(maxuv, maxpq) if(is.null(h.start)) h.start = max.order +1 if(is.null(llh.start)) llh.start =1 if(init.rec !="mci"& model.var !="garch"){ stop("Algorithm only supported for mci Recursion") } ans = list(model = c(model.mean, model.var), order = c(u = u, v = v, p = p, q = q), max.order = max.order, z = rep(0, times = length(series)), #这里的0,长度为444 h = rep(var(series), times = length(series)), #h是方差是1(因为此时的series是被标准化过的),长度为444 x = series, scale = scale, init.rec = init.rec, h.start = h.start, llh.start = llh.start) ans}


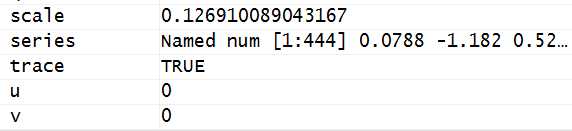
.params <-.garchInitParameters(formula.mean = formula.mean, formula.var = formula.var, delta = delta, skew = skew, shape = shape, cond.dist = cond.dist[1], include.mean = include.mean, include.delta = include.delta, include.skew = include.skew, include.shape = include.shape, leverage = leverage, algorithm = algorithm[1], control = con, trace = trace) .setfGarchEnv(.params =.params)function (formula.mean, formula.var, delta, skew, shape, cond.dist, include.mean, include.delta, include.skew, include.shape, leverage, algorithm, control, trace) { .DEBUG = FALSE .series <-.getfGarchEnv(".series") model.order =as.numeric(strsplit(strsplit(strsplit(as.character(formula.mean), "\\(")[[2]][2],"\\)")[[1]],",")[[1]]) u = model.order[1] v =0 if(length(model.order)==2) v = model.order[2] model.order =as.numeric(strsplit(strsplit(strsplit(as.character(formula.var), "\\(")[[2]][2],"\\)")[[1]],",")[[1]]) p = model.order[1] q =0 if(length(model.order)==2) q = model.order[2] model.var =.series$model[2] if(is.null(include.delta)){ if(model.var =="garch"){ include.delta = FALSE } else{ include.delta = TRUE } } if(is.null(leverage)){ if(model.var =="garch"){ leverage = FALSE } else{ leverage = TRUE } } if(cond.dist =="t") cond.dist ="std" skewed.dists = c("snorm","sged","sstd","snig") if(is.null(include.skew)){ if(any(skewed.dists == cond.dist)){ include.skew = TRUE } else{ include.skew = FALSE } } shaped.dists = c("ged","sged","std","sstd","snig") if(is.null(include.shape)){ if(any(shaped.dists == cond.dist)){ include.shape = TRUE } else{ include.shape = FALSE } } Names= c("mu", if(u >0) paste("ar",1:u, sep =""), if(v >0) paste("ma",1:v, sep =""),"omega", if(p >0) paste("alpha",1:p, sep =""), if(p >0) paste("gamma",1:p, sep =""), if(q >0) paste("beta",1:q, sep =""), "delta","skew","shape") fit.mean = arima(.series$x, order = c(u,0, v), include.mean = include.mean)$coef alpha.start =0.1 beta.start =0.8 params = c( if(include.mean) fit.mean[length(fit.mean)]else0, #最后一项就是intercept,所以就是mu啊! if(u >0) fit.mean[1:u], if(v >0) fit.mean[(u +1):(length(fit.mean)-as.integer(include.mean))], var(.series$x, na.rm = TRUE)*(1- alpha.start - beta.start), if(p >0) rep(alpha.start/p, times = p), if(p >0) rep(0.1, times = p), if(q >0) rep(beta.start/q, times = q), delta, skew, shape) names(params)=Names TINY =1e-08 USKEW =1/10 USHAPE =1 if(cond.dist =="snig") USKEW =-0.99 U = c(-10* abs(mean(.series$x)),if(u >0) rep(-1+ TINY, times = u),if(v >0) rep(-1+ TINY, times = v),1e-06* var(.series$x),if(p >0) rep(0+ TINY, times = p), if(p >0) rep(-1+ TINY, times = p),if(q >0) rep(0+ TINY, times = q),0, USKEW, USHAPE) names(U)=Names VSKEW =10 VSHAPE =10 if(cond.dist =="snig") VSKEW =0.99 V = c(10* abs(mean(.series$x)),if(u >0) rep(1- TINY, times = u),if(v >0) rep(1- TINY, times = v),100* var(.series$x),if(p >0) rep(1- TINY, times = p), if(p >0) rep(1- TINY, times = p),if(q >0) rep(1- TINY, times = q),2, VSKEW, VSHAPE) names(V)=Names includes = c(include.mean,if(u >0) rep(TRUE, times = u), if(v >0) rep(TRUE, times = v), TRUE,if(p >0) rep(TRUE, times = p),if(p >0) rep(leverage, times = p), if(q >0) rep(TRUE, times = q), include.delta, include.skew, include.shape) names(includes)=Names index =(1:length(params))[includes == TRUE] names(index)= names(params)[includes == TRUE] alpha <- beta <- NULL if(p >0) alpha = params[substr(Names,1,5)=="alpha"] if(p >0& leverage) gamma = params[substr(Names,1,5)=="gamma"] if(p >0&!leverage) gamma = rep(0, times = p) if(q >0) beta = params[substr(Names,1,4)=="beta"] if(.series$model[2]=="garch"){ persistence = sum(alpha)+ sum(beta) } elseif(.series$model[2]=="aparch"){ persistence = sum(beta) for(i in1:p) persistence = persistence + alpha[i]* garchKappa(cond.dist, gamma[i], params["delta"], params["skew"], params["shape"]) } names(persistence)="persistence" list(params = params, U = U, V = V, includes = includes, index = index, mu = params[1], delta = delta, skew = skew, shape = shape, cond.dist = cond.dist, leverage = leverage, persistence = persistence, control = control)}fit.mean = arima(.series$x, order = c(u,0, v), include.mean = include.mean)$coefparams = c( if(include.mean) fit.mean[length(fit.mean)]else0{.series <-.getfGarchEnv(".series").params <-.getfGarchEnv(".params") INDEX =.params$index algorithm =.params$control$algorithm[1] #algorithm是nlminb TOL1 =.params$control$tol1 TOL2 =.params$control$tol2if(algorithm =="nlminb"| algorithm =="nlminb+nm"){ fit <-.garchRnlminb(.params,.series,.garchLLH, trace).params$llh = fit$llh.params$params[INDEX]= fit$par.setfGarchEnv(.params =.params)}.params$llh = fit$llh.params$params[INDEX]= fit$par.setfGarchEnv(.params =.params)if(hessian =="ropt"){ fit$hessian <--.garchRoptimhess(par = fit$par,.params =.params,.series =.series) titleHessian ="R-optimhess"}...............if(.params$control$xscale){.series$x <-.series$x *.series$scale #将标准化后的值还原为原始的数值了if(.params$include["mu"]) fit$coef["mu"]<- fit$par["mu"]<-.params$params["mu"]<-.params$params["mu"]*.series$scaleif(.params$include["omega"]) fit$coef["omega"]<- fit$par["omega"]<-.params$params["omega"]<-.params$params["omega"]*.series$scale^(.params$params["delta"]).setfGarchEnv(.params =.params).setfGarchEnv(.series =.series)}if(.params$control$xscale){if(.params$include["mu"]){ fit$hessian[,"mu"]<- fit$hessian[,"mu"]/.series$scale fit$hessian["mu",]<- fit$hessian["mu",]/.series$scale}if(.params$include["omega"]){ fit$hessian[,"omega"]<- fit$hessian[,"omega"]/.series$scale^(.params$params["delta"]) fit$hessian["omega",]<- fit$hessian["omega",]/.series$scale^(.params$params["delta"])}}.llh <- fit$llh <- fit$value <-.garchLLH(fit$par, trace = FALSE, fGarchEnv = TRUE).series <-.getfGarchEnv(".series")if(robust.cvar) fit$gradient <--.garchRCDAGradient(par = fit$par,.params =.params,.series =.series) N = length(.series$x) NPAR = length(fit$par) fit$ics = c(AIC = c((2* fit$value)/N +2* NPAR/N), BIC =(2* fit$value)/N + NPAR * log(N)/N, SIC =(2* fit$value)/N + log((N +2* NPAR)/N), HQIC =(2* fit$value)/N +(2* NPAR * log(log(N)))/N) names(fit$ics)<- c("AIC","BIC","SIC","HQIC") fit}function (.params,.series,.garchLLH, trace) { if(trace) cat("\nR coded nlminb Solver: \n\n") INDEX =.params$index parscale = rep(1, length = length(INDEX)) names(parscale)= names(.params$params[INDEX]) parscale["omega"]= var(.series$x)^(.params$delta/2) parscale["mu"]= abs(mean(.series$x)) #这个并非mu的初始值 TOL1 =.params$control$tol1 fit <- nlminb(start =.params$params[INDEX], objective =.garchLLH, lower =.params$U[INDEX], upper =.params$V[INDEX], scale =1/parscale, control = list(eval.max =2000, iter.max =1500, rel.tol =1e-14* TOL1, x.tol =1e-14* TOL1, trace =as.integer(trace)), fGarchEnv = FALSE) fit$value <- fit.llh <- fit$objective names(fit$par)= names(.params$params[INDEX]) .garchLLH(fit$par, trace = FALSE, fGarchEnv = TRUE) fit$coef <- fit$par fit$llh <- fit$objective fit}fit$coef["mu"]<- fit$par["mu"]<-.params$params["mu"]<-.params$params["mu"]* .series$scale.garchLLHfunction (params, trace = TRUE, fGarchEnv = FALSE) { DEBUG = FALSE if(DEBUG) print("Entering Function .garchLLH") .series <-.getfGarchEnv(".series") .params <-.getfGarchEnv(".params") .garchDist <-.getfGarchEnv(".garchDist") #概率密度函数 .llh <-.getfGarchEnv(".llh") #对数似然函数 if(DEBUG) print(.params$control$llh) if(.params$control$llh =="internal"){ if(DEBUG) print("internal") return(.aparchLLH.internal(params, trace = trace, fGarchEnv = fGarchEnv)) #结束 } elseif(.params$control$llh =="filter"){ if(DEBUG) print("filter") return(.aparchLLH.filter(params, trace = trace, fGarchEnv = fGarchEnv)) } elseif(.params$control$llh =="testing"){ if(DEBUG) print("testing") return(.aparchLLH.testing(params, trace = trace, fGarchEnv = fGarchEnv)) } else{ stop("LLH is neither internal, testing, nor filter!") }}<environment: namespace:fGarch>function (params, trace = TRUE, fGarchEnv = TRUE) { DEBUG = FALSE if(DEBUG) print("Entering Function .garchLLH.internal") .series <-.getfGarchEnv(".series") .params <-.getfGarchEnv(".params") .garchDist <-.getfGarchEnv(".garchDist") .llh <-.getfGarchEnv(".llh") if(DEBUG) print(.params$control$llh) if(.params$control$llh =="internal"){ INDEX <-.params$index MDIST <- c(norm =10, QMLE =10, snorm =11, std =20, sstd =21, ged =30, sged =31)[.params$cond.dist] if(.params$control$fscale) NORM <- length(.series$x) else NORM =1 REC <-1 if(.series$init.rec =="uev") REC <-2 MYPAR <- c(REC = REC, LEV =as.integer(.params$leverage), MEAN =as.integer(.params$includes["mu"]), DELTA =as.integer(.params$includes["delta"]), SKEW =as.integer(.params$includes["skew"]), SHAPE =as.integer(.params$includes["shape"]), ORDER =.series$order, NORM =as.integer(NORM)) MAX <- max(.series$order) NF <- length(INDEX) N <- length(.series$x) DPARM <- c(.params$delta,.params$skew,.params$shape) fit <-.Fortran("garchllh", N =as.integer(N), Y =as.double(.series$x), Z =as.double(.series$z), H =as.double(.series$h), NF =as.integer(NF), X =as.double(params), DPARM =as.double(DPARM), MDIST =as.integer(MDIST), MYPAR =as.integer(MYPAR), F =as.double(0), PACKAGE ="fGarch") llh <- fit[[10]] #此时llh就是-312 if(is.na(llh)) llh =.llh +0.1*(abs(.llh)) if(!is.finite(llh)) llh =.llh +0.1*(abs(.llh)) .setfGarchEnv(.llh = llh) if(fGarchEnv){ .series$h <- fit[[4]] .series$z <- fit[[3]] #直到这里,z才从444个0被替换为残差 .setfGarchEnv(.series =.series) } } else{ stop("LLH is not internal!") } c(LogLikelihood= llh)}Browse[5]>.garchDistfunction (z, hh, skew, shape) { dnorm(x = z/hh, mean =0, sd =1)/hh}<environment: namespace:fGarch>使用RStudio调试(debug)基础学习(二)和fGarch包中的garchFit函数估计GARCH模型的原理和源码
原文:http://www.cnblogs.com/xuanlvshu/p/6354091.html